A segmentation guide and probabilistic atlas of the C57BL/6J mouse brain from magnetic resonance imaging
Jeremy Ullmann (The University of Queensland), Charles Watson (Curtin University), Andrew Janke (The University of Queensland), Marianne Keller (The University of Queensland), Nyoman Kurniawan (The University of Queensland), Zhengyi Yang (The University of Queensland), Kay Richards (Florey Neurosciences Institute), George Paxinos (The University of New South Wales, ), Gary Egan (Monash University), Steven Petrou (Florey Neuroscience Institutes), Perry Bartlett (The University of Queensland), Graham Galloway (The University of Queensland), David Reutens (The University of Queensland)
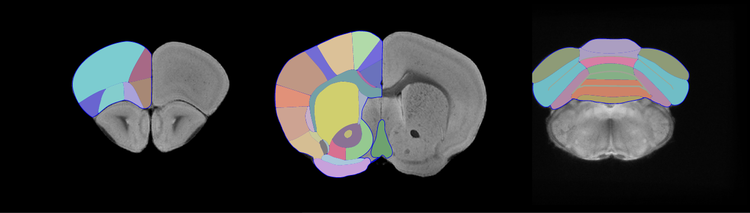
MRI data from 18 mice was acquired as per Janke et al.3. Images were placed in the stereotaxic Waxholm space3 and a symmetric model was created4. The components of the brain were then delineated, on the bases of differences in signal intensity and/or their location in reference to landmark structures, and partitioned using vector-based segmentation via a Cintiq tablet.
We acquired 30µm3 data sets, which were averaged to create a model resolution of 15µm3. We developed a detailed parcellation scheme for segmenting the C57BL/6J mouse brain. It is based on MRI landmarks, which are reproducible, and visible as a consequence of the higher signal-to-noise ratio achieved during group averaging and the inter-subject stability of structures. The segmentation protocol creates standardized structural delineations that can be applied to studies examining mouse models of neurological disease. We also generated a digital atlas containing over 200 structures with mean region volumes, T2*-weighted signal intensities and probability maps for each of structure. This atlas offers a detailed template for cross modality applications and is online at www.imaging.org.au/AMBMC.
1 Dorr, et al. Neuroimage 42, 60-69 (2008).2 Franklin, K. & Paxinos, G. The mouse brain in stereotaxic coordinates. 3 edn, (Academic Press, 2008).3 Johnson, et al. Waxholm Space: Neuroimage 53, 365-372 (2010).4 Janke et al. 15um average mouse models in Waxholm space from 16.4T 30um images. In 20th Annual ISMRM Scientific Meeting and Exhibition, Melbourne, Australia (2012).

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
