A Semi-automated Program for Axonal Reconstructions from Time-lapse 2-Photon Images
Zhilun Yang (School of Software, Tsinghua University), Qian Wang (Department of Biomedical Engineering, School of Medicine, Tsinghua University), Ge Gao (Department of Biomedical Engineering, School of Medicine, Tsinghua University), Xuesong Li (Department of Biomedical Engineering, School of Medicine, Tsinghua University), Federico Grillo (MRC Clinical Sciences Centre, Imperial College London, Du Cane road, London), Valentina Ferretti (MRC Clinical Sciences Centre, Imperial College London, Du Cane road, London), Vincenzo De Paola (MRC Clinical Sciences Centre, Imperial College London, Du Cane road, London), Sen Song (Department of Biomedical Engineering, School of Medicine, Tsinghua University)
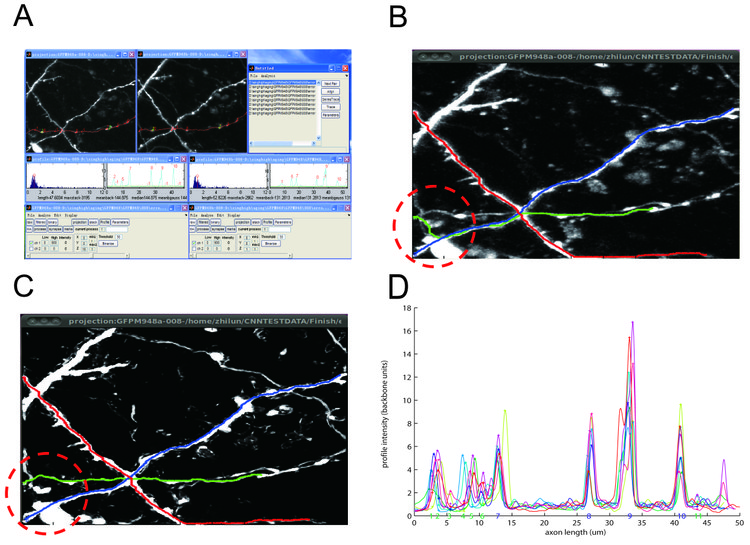
We use two-photon time-lapse images of GFP-expressing axons from mouse barrel cortex taken in the De Paola lab. Firstly, we use median filtering to get rid of shot noise (figure B). To get rid of remaining unwanted background fluorescence and resolve individual neurites, we follow a recent machine learning approach proposed by the Sebastian Seung lab (Sumbul U et al, Neuroscience Abstracts 2011) and use a convolutional neural network for enhancement (figure C). Secondly, we incorporated the algorithm from the Simple Neuron Tracer Fiji plugin to automatically trace a path between two points (Figure B and C). The result shows that the convolutional neural network is able to help resolve junction points and reduce unspecific background. The much cleaner image led to a dramatic improvement in tracing speed (2.92±0.29s versus 15.7±4.7s for each axon), making the user experience more efficient. In some instances, tracing on the median filtered image gave the wrong result due to axons coming close to each other while the result from the enhanced image is correct (figure B versus C). Thirdly, we extract the axonal backbone intensity and align it over imaging sessions using fiducial points (figure D ). Lastly, we use local intensity peak finding to extract bouton strength values. We are currently using this software to analyze the synaptic strength changes underlying cortical remapping. Future improvements in over-day alignment are expected to further reduce the amount of user efforts. We plan to release NeuronMatrix to the general community as a Fiji plugin in the future.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
