A spatial coding scheme to define the neuron types in the Drosophila brain
Filed under:
Computational neuroscience
Chao-Chun Chuang (National Center for High-Performance Computing; Institute of Bioinformatics, National Chiao Tung University), Chang-Wei Yeh (National Center for High-Performance Computing), Chang-Huain Hsieh (National Center for High-Performance Computing), Hsiu-Ming Chang (Brain Research Center, National Tsing Hua University), Jenn-Kang Hwang (Institute of Bioinformatics and Systems Biology, National Chiao Tung University)
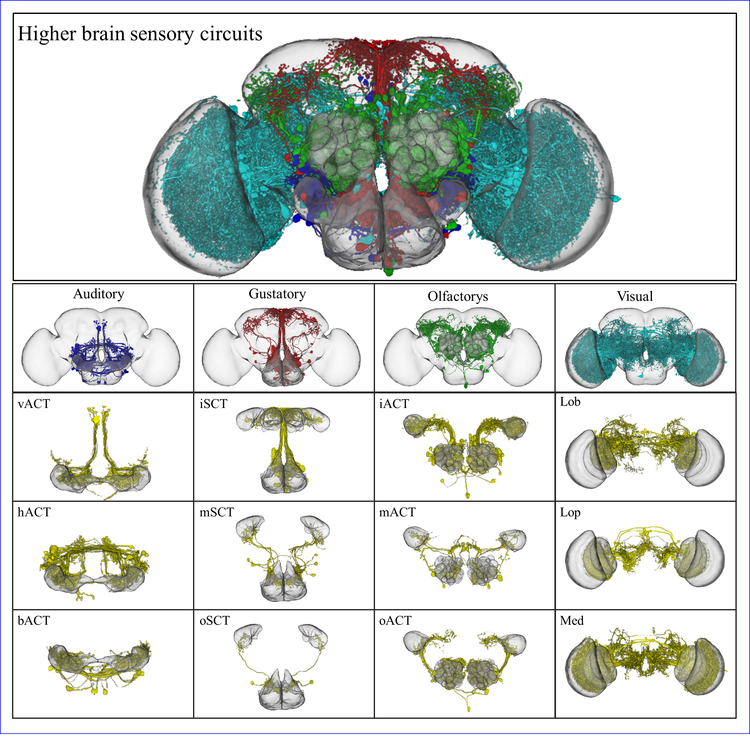
A neuron type is an element to be manipulated for neuronal circuit analysis in the Drosophila brain. With classification and definition of neuron types, we will have a detailed look at how neurons work together. In this report, we provide a method to classify neuron types in the Drosophila brain from their spatial distributions in the brain: neurons exhibit the similar structure in the circuit belongs to the same neuron type.
However, massive morphology alignment with direct 3D structural comparison is too difficult to be carried out in a high-throughput screening procedure. Thus, we developed a new approach to translate 3D neuronal morphology into the one-dimensional spatial sequence. By the alignment of the spatial sequences, we can find the neurons with similar morphology disregard their lateralization and gender differences. Finally, we have clustered 689 AL PNs in Flycircuit in to 76 neuron types and found that projection neurons with very similar morphology were formed at different developmental stages. Other cases, we found multiple genes were expressed neurons of similar morphology. In addition, sexual dimorphisms in neuron structures were detected. These results corresponded with the actual anatomy atlas, demonstrating our algorithm to be effective and accurate in a high-throughput screening procedure.
In conclusion, we provide a novel approach to integrate anatomy and informatics. It can handle massive 3D neuronal image data collected in experiments from different research groups as well as manage bio-images with deeper neurological insight.
However, massive morphology alignment with direct 3D structural comparison is too difficult to be carried out in a high-throughput screening procedure. Thus, we developed a new approach to translate 3D neuronal morphology into the one-dimensional spatial sequence. By the alignment of the spatial sequences, we can find the neurons with similar morphology disregard their lateralization and gender differences. Finally, we have clustered 689 AL PNs in Flycircuit in to 76 neuron types and found that projection neurons with very similar morphology were formed at different developmental stages. Other cases, we found multiple genes were expressed neurons of similar morphology. In addition, sexual dimorphisms in neuron structures were detected. These results corresponded with the actual anatomy atlas, demonstrating our algorithm to be effective and accurate in a high-throughput screening procedure.
In conclusion, we provide a novel approach to integrate anatomy and informatics. It can handle massive 3D neuronal image data collected in experiments from different research groups as well as manage bio-images with deeper neurological insight.
Preferred presentation format:
Poster
Topic:
Computational neuroscience

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
