CAJAL3D: Towards A Fully Automatic Pipeline for Connectome Estimation from High-Resolution EM Data
Dean M. Kleissas (Johns Hopkins University, Applied Physics Laboratory), William R. Gray (Johns Hopkins University, Applied Physics Laboratory; Johns Hopkins University), James M. Burck (Johns Hopkins University, Applied Physics Laboratory), Joshua T. Vogelstein (Johns Hopkins University), Eric Perlman (Janelia Farm Research Campus, HHMI), Philippe M. Burlina (Johns Hopkins University, Applied Physics Laboratory), Randal Burns (Johns Hopkins University), R. Jacob Vogelstein (Johns Hopkins University, Applied Physics Laboratory; Johns Hopkins University)
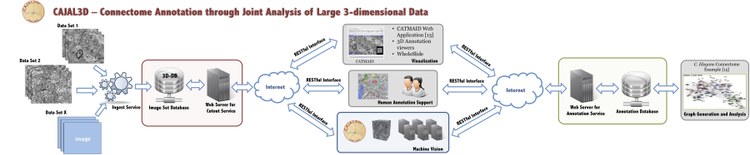
In addition to defining a common interface for connectomics, we developed a processing pipeline framework that implements our annotation standard and is integrated with the Open Connectome Project (OCP) web services. The framework is built on a client-server based infrastructure that facilitates scalable, distributed processing. The pipeline exchanges data and data products associated with various forms of connectomics information, ranging from raw images to processed graphs, with the OCP storage servers. The annotation standard, image processing algorithms, image and annotation storage databases and associated web services are being made available to the community as a free resource. We are in the process of soliciting feedback from the community for additional features and functionality.
DMK and WRG contributed equally to this work.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
