An indirect encoding scheme for artificial neural networks based on gene regulatory networks
Filed under:
Computational neuroscience
Martin Pyka (Department of Psychiatry, University of Marburg), Sascha Hauke (Telecooperation Group, Technische Universität Darmstadt), Dominik Heider (Department of Bioinformatics, University of Duisburg-Essen)
The architecture of natural neural networks evolved through a gradual complexification driven by evolutionary pressure and generated through a highly indirect encoding scheme. Although the development of organic structures (such as neural networks) from the genotype happens through local interactions of proteins and cells, global coordinated structures and patterns can be observed in the phenotype.
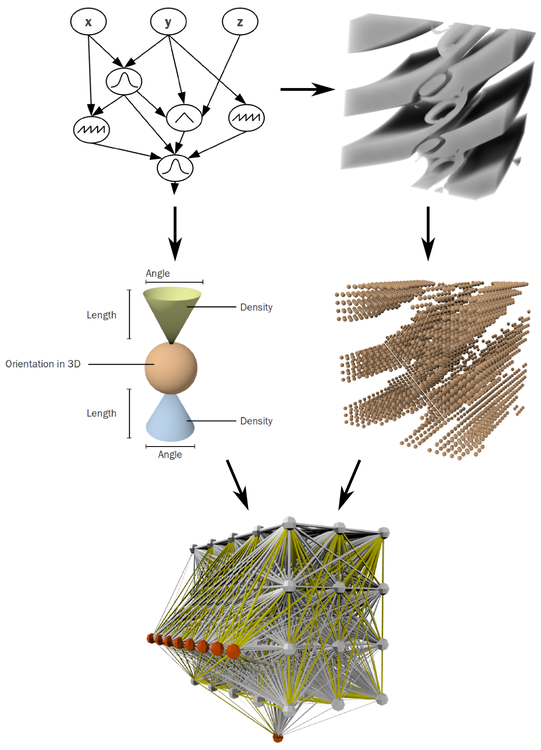
A Compositional Pattern Producing Network (CPPN) is a model for building such patterns using principles of gene regulatory networks, while exploiting shortcuts in the simulated world (Stanley, 2007). The main idea behind CPPNs is that single genes are activated at a certain concentration level of proteins generated by other genes. Thus, the concentration gradient of a gene can be regarded as the activation function of another protein/gene concentration. Given that different genes have different activation functions and differing influences on other genes, pattern formation can be represented as a graph of concatenated activation functions. The output of the last function builds the pattern for a given substrate. CPPNs are a highly indirect encoding scheme. However, the phenotype is stable and robust against continues changes in the genotype, making it interesting for evolution-driven approaches.
We present a model based on CPPNs, called Brain-in-a-box (BIB), that implements several mechanisms of natural developmental processes. Neurons are placed in space according to patterns generated by a BIB, potential connections between neurons are derived from axon- and dendrite cones whose properties are also controlled by the BIB. Thus, neural networks for controlling organisms can be generated and improved through evolutionary methods with indirect genotype-phenotype mechanisms, as they occur in biological systems.
Several examples of this model are presented, demonstrating its functionality and how it can be extended to incorporate concepts, such as synaptic plasticity and neuromodulation. The framework and the demos are implemented in Python using the Briansimulator (Goodman and Brette, 2008).
Goodman D and Brette R (2008) Brian: a simulator for spiking neural networks in Python. Front. Neuroinform. http://dx.doi.org/10.3389/neuro.11.005.2008
Stanley K (2007) Compositional pattern producing networks: A novel abstraction of development. Genetic Programming and Evolvable Machines, 8(2):131–162
A Compositional Pattern Producing Network (CPPN) is a model for building such patterns using principles of gene regulatory networks, while exploiting shortcuts in the simulated world (Stanley, 2007). The main idea behind CPPNs is that single genes are activated at a certain concentration level of proteins generated by other genes. Thus, the concentration gradient of a gene can be regarded as the activation function of another protein/gene concentration. Given that different genes have different activation functions and differing influences on other genes, pattern formation can be represented as a graph of concatenated activation functions. The output of the last function builds the pattern for a given substrate. CPPNs are a highly indirect encoding scheme. However, the phenotype is stable and robust against continues changes in the genotype, making it interesting for evolution-driven approaches.
We present a model based on CPPNs, called Brain-in-a-box (BIB), that implements several mechanisms of natural developmental processes. Neurons are placed in space according to patterns generated by a BIB, potential connections between neurons are derived from axon- and dendrite cones whose properties are also controlled by the BIB. Thus, neural networks for controlling organisms can be generated and improved through evolutionary methods with indirect genotype-phenotype mechanisms, as they occur in biological systems.
Several examples of this model are presented, demonstrating its functionality and how it can be extended to incorporate concepts, such as synaptic plasticity and neuromodulation. The framework and the demos are implemented in Python using the Briansimulator (Goodman and Brette, 2008).
Goodman D and Brette R (2008) Brian: a simulator for spiking neural networks in Python. Front. Neuroinform. http://dx.doi.org/10.3389/neuro.11.005.2008
Stanley K (2007) Compositional pattern producing networks: A novel abstraction of development. Genetic Programming and Evolvable Machines, 8(2):131–162
Preferred presentation format:
Poster
Topic:
Computational neuroscience

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
