Electrophysiology lab automation: a case study
Filed under:
Electrophysiology
Rembrandt Bakker (Donders Institute for Brain, Cognition and Behaviour, Radboud University Nijmegen), Dirk Schubert (Donders Institute for Brain, Cognition and Behaviour, Radboud University Nijmegen Medical Centre), Paul Tiesinga (Donders Institute for Brain, Cognition and Behaviour, Radboud University Nijmegen)
A major hurdle for sharing neuroscientific data is the effort to annotate it with meta data: the parameters that describe the circumstances relevant to the recorded data. Traditionally, meta data is distributed over a lab journal (parameters that change often) and the experimenter's personal memory (constant parameters). To get the meta data in electronic form, one needs to design an ontology that covers all parameters that are relevant to the experiments, and then extract these parameters from the lab journal and personal memory. We present a case study from an electrophysiology lab to show that carrying out these steps is a rewarding investment that not only enables data sharing but also the construction of automated analysis pipelines.
Our database concerns in vitro Multi Electrode Array (MEA) and single cell patch clamp measurements from the rat barrel cortex. The meta data ontology is implemented as a relational database scheme, available at http://mealab.science.ru.nl/main/scheme.php. We created a MySQL database according to this scheme, and populated it with thousands of MEA experiments. For the analysis pipeline, we rely on three associated web services:
(1) A meta data search wizard (http://mealab.science.ru.nl/main/search_wizard.php) to find experiments that meet certain criteria; the available choices are inferred from the database scheme.
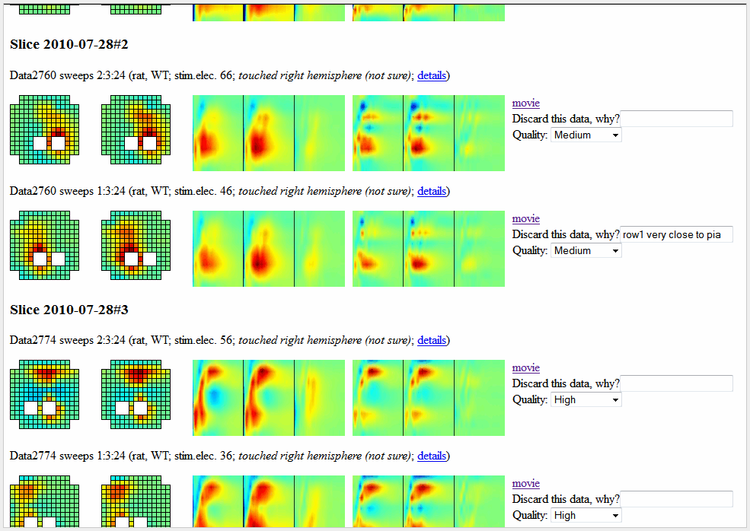
(2) An interactive webpage that groups and displays key properties and images of recordings, with links to detailed info (see figure). Each entry contains a field 'Quality', where one can assign a quality rating to the experiment.
(3) An SQL query service that returns tables in JSON format. This allows web-based access to the database without the need to install drivers.
The automated analysis is implemented in Matlab. The actual recordings are stored on local hard drives. We hope that publication of the database scheme and services will foster the development of general purpose lab automation systems (e.g., Grewe, Wachtler and Benda 2011, doi: 10.3389/fninf.2011.00016) and the specification of meta data standards. Our research aim is to study stimulus-response patterns in slices with different genotypes or farmacological treatments. Direct benefits of the database are: (1) a new research question can be answered by modifying a single SQL condition; (2) old experiments are not forgotten; and (3) base-line statistics are much improved by combining all experiments that start with similar conditions.
Our database concerns in vitro Multi Electrode Array (MEA) and single cell patch clamp measurements from the rat barrel cortex. The meta data ontology is implemented as a relational database scheme, available at http://mealab.science.ru.nl/main/scheme.php. We created a MySQL database according to this scheme, and populated it with thousands of MEA experiments. For the analysis pipeline, we rely on three associated web services:
(1) A meta data search wizard (http://mealab.science.ru.nl/main/search_wizard.php) to find experiments that meet certain criteria; the available choices are inferred from the database scheme.
(2) An interactive webpage that groups and displays key properties and images of recordings, with links to detailed info (see figure). Each entry contains a field 'Quality', where one can assign a quality rating to the experiment.
(3) An SQL query service that returns tables in JSON format. This allows web-based access to the database without the need to install drivers.
The automated analysis is implemented in Matlab. The actual recordings are stored on local hard drives. We hope that publication of the database scheme and services will foster the development of general purpose lab automation systems (e.g., Grewe, Wachtler and Benda 2011, doi: 10.3389/fninf.2011.00016) and the specification of meta data standards. Our research aim is to study stimulus-response patterns in slices with different genotypes or farmacological treatments. Direct benefits of the database are: (1) a new research question can be answered by modifying a single SQL condition; (2) old experiments are not forgotten; and (3) base-line statistics are much improved by combining all experiments that start with similar conditions.
Preferred presentation format:
Poster
Topic:
Electrophysiology

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
