First generation Waxholm atlas of the Sprague-Dawley rat brain
Filed under:
Digital atlasing
Eszter A. Papp (CMBN, Institute of Basic Medical Sciences, University of Oslo), Trygve B. Leergaard (CMBN, Institute of Basic Medical Sciences, University of Oslo), G. Allan Johnson (Center for in vivo Microscopy, Dept Radiology, Duke University Medical Center), Jan G. Bjaalie (CMBN, Institute of Basic Medical Sciences, University of Oslo)
Standardized brain atlas spaces provide key anchoring points for comparing heterogeneous datasets from different experimental animals in the context of structure - function analysis. The usability of such an atlas space depends on common access to high quality reference material, including delineations of anatomical structures (labels), and underlying original images (templates). Analysis of microscopic data of 3D nature, e.g. gene expression distributions and connectivity patterns, can to advantage make use of high resolution non-distorted volumetric templates and associated atlas labels for the rat brain. We present a first generation of such reference material for the standard Waxholm Space in the Sprague-Dawley rat.
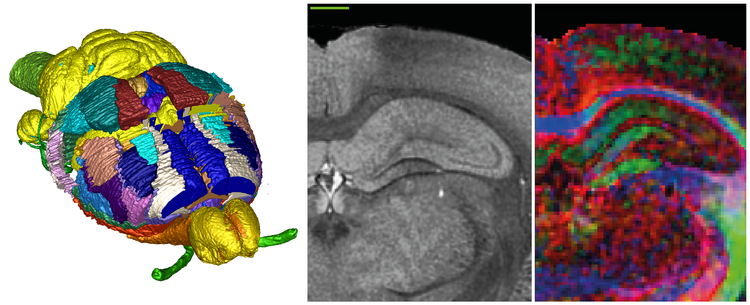
Microscopic resolution ex vivo magnetic resonance images (MRI) were acquired from an 80 day old male Sprague-Dawley rat, including T2* anatomical images with 39 μm isotropic voxels, and diffusion tensor images (DTI) with 78 μm isotropic voxels. Anatomical structures, including nuclei, areas, and fiber tracts, were delineated based on image contrast using ITK-SNAP software, resulting in 67 distinct labels along with detailed delineation criteria. Labels representing cortical areas have been transferred from an existing atlas (Hjornevik et al., http://dx.doi.org/10.3389/neuro.11.004.2007), warped into the volumetric template, and manually adjusted for shape differences. Validation of atlas labels was aided by collections of histological images from brains of comparable animals, and delineations from other rat brain atlases.
The volumetric template, as well as the atlas labels, will be made open access through the INCF Software Center. Spatial reference is provided by the application of Waxholm Space, a standard atlas space recently defined by the International Neuroinformatics Coordinating Facility (INCF). The use of Waxholm Space connects the atlas to a growing infrastructure of interoperable resources and services for multi-level data integration and analysis across reference spaces.
The presented atlas serves as a basis for a library of individual volumetric templates for different applications, e.g. different strains, disease models, and developmental stages. The atlas is to be included in a server-based registration pipeline as part of the INCF Digital Atlasing Infrastructure, allowing registration of both experimental data and new suitable templates to the atlas.
Microscopic resolution ex vivo magnetic resonance images (MRI) were acquired from an 80 day old male Sprague-Dawley rat, including T2* anatomical images with 39 μm isotropic voxels, and diffusion tensor images (DTI) with 78 μm isotropic voxels. Anatomical structures, including nuclei, areas, and fiber tracts, were delineated based on image contrast using ITK-SNAP software, resulting in 67 distinct labels along with detailed delineation criteria. Labels representing cortical areas have been transferred from an existing atlas (Hjornevik et al., http://dx.doi.org/10.3389/neuro.11.004.2007), warped into the volumetric template, and manually adjusted for shape differences. Validation of atlas labels was aided by collections of histological images from brains of comparable animals, and delineations from other rat brain atlases.
The volumetric template, as well as the atlas labels, will be made open access through the INCF Software Center. Spatial reference is provided by the application of Waxholm Space, a standard atlas space recently defined by the International Neuroinformatics Coordinating Facility (INCF). The use of Waxholm Space connects the atlas to a growing infrastructure of interoperable resources and services for multi-level data integration and analysis across reference spaces.
The presented atlas serves as a basis for a library of individual volumetric templates for different applications, e.g. different strains, disease models, and developmental stages. The atlas is to be included in a server-based registration pipeline as part of the INCF Digital Atlasing Infrastructure, allowing registration of both experimental data and new suitable templates to the atlas.
Preferred presentation format:
Poster
Topic:
Digital atlasing

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
