Integrated Analysis of Anatomical Gene Expression Maps and Co-Expression Networks Using a Database, ViBrism
Yuko Okamura-Oho (BReNt-Brain Research Network and Advanced Science Institute, RIKEN), Kazuro Shimokawa (Department of Gene Diagnostics and Therapeutics, Research Institute, National Center for Global Health and Medicine), Satoko Takemoto (Bio-research Infrastructure Construction Team, Advanced Science Institute, RIKEN ), Gang Song (Penn Image Computing and Science Laboratory, University of Pennsylvania), James Gee (Penn Image Computing and Science Laboratory, University of Pennsylvania), Hideo Yokota (Bio-research Infrastructure Construction Team, Advanced Science Institute, RIKEN )
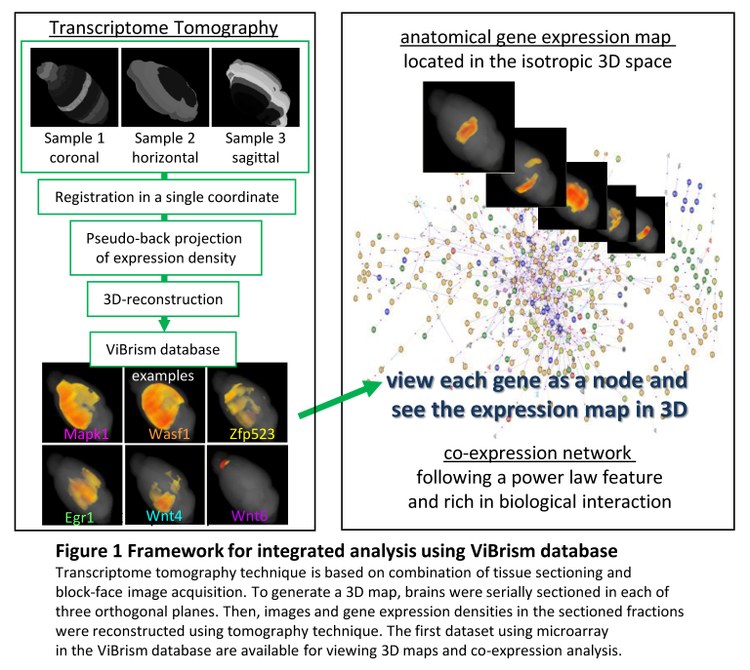
To determine the association systematically, we have introduced a novel framework, Transcriptome Tomography, for spatially integrating comprehensive endogenous gene expression within an isotropic anatomical context. Using this rapid and unbiased 3D mapping technique, in the first instance, we have generated a dataset of 36,000 maps covering the whole mouse brain (ViBrism: http://vibrism.riken.jp/3dviewer/ex/index.html) and validated them against existing data with respect to the expression location and density (paper submitted).
Here, we used an informatics approach to identify the combinatorial gene expression as a broad co-expression network. The gene network links covering the whole brain followed an inverse-power law and were rich in functional interaction and gene ontology terms. Developmentally conserved co-expression modules underlie the network structure. To demonstrate the relevance of the finding, we mined Huntington’s disease gene (Htt) and found a novel disease-related co-expression network containing genes potentially co-functioning with Htt in neural differentiation and modulating the disease specific differential vulnerability in brain regions.
The maps are spatially isotropic and well suited to analysis in the standard space for brain-atlas databases, e.g. Waxholm Space (PLoS Comput Biol 2011, 7[2]: e1001065) as shown in the related poster by J. Boline et., al. Our time and cost effective framework will facilitate research creating and using open-resources for a molecular-based understanding of complex structures.
A part of this work was conducted within the Waxholm Space Task Force of the International Neuroinformatics Coordinating Facility (INCF) Program on Digital Brain Atlasing. We thank the program members, particularly, R. Baldock, I. Zaslavsky, L.Ibanez and J. Boline.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
