Intuitive and efficient deployment of neuroimaging pipelines in clinical research with BRICpipe
Filed under:
Clinical neuroscience
Andreas Glatz (BRIC, Division of Clinical Neurosciences, University of Edinburgh), Colin R. Buchanan (School of Informatics, University of Edinburgh), Maria C. Valdés Hernández (BRIC, Division of Clinical Neurosciences, University of Edinburgh), Mark E. Bastin (BRIC, Division of Clinical Neurosciences, University of Edinburgh), Joanna M. Wardlaw (BRIC, Division of Clinical Neurosciences, University of Edinburgh)
INTRODUCTION
In clinical research, neuroimaging pipelines are mainly used to combine the strength of single analysis tools and to accelerate the data analysis. Pipeline frameworks, such as Nipype[1] or LONI[2] (Table 1), should be a researcher’s first choice to implement pipelines. However, clinical researchers often prefer Bash[3] scripts due to their simplicity and because pipeline frameworks still require specialist knowledge for setup and use.
To encourage the use of pipeline frameworks in clinical research, we developed our own open source pipeline framework, BRICpipe. Here, we present BRICpipe’s design and some validation results to show how BRICpipe compares to Nipype and Bash scripts.
DESIGN
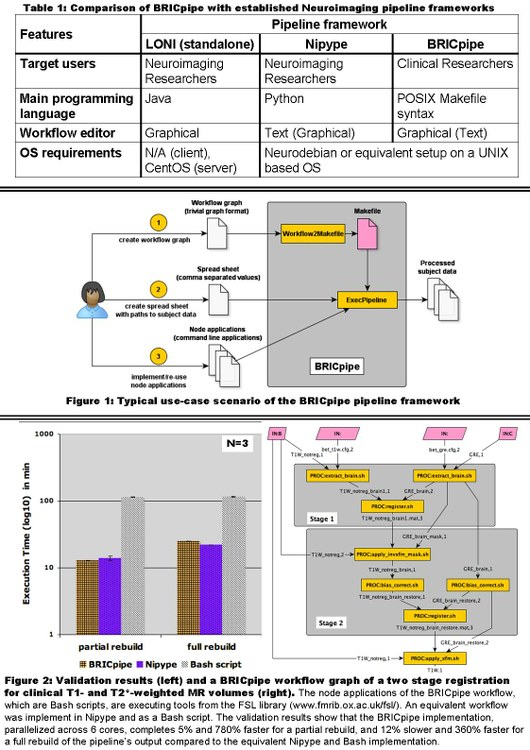
Figure 1 shows the typical use-case of BRICpipe: the user has to supply a workflow graph consisting of input, output and processing nodes (e.g. Figure 2), a spreadsheet with paths to input data from all subjects, and command line applications for each processing node. The workflow graph is designed in a graph editor (e.g. yEd[4]) and converted to a Makefile with the Workflow2Makefile application. Lastly, the ExecWorkflow application starts the pipeline execution by invoking GNU make[3] as execution engine.
VALIDATION
We validated BRICpipe with a two stage registration pipeline (Figure 2) for clinical T1- and T2*-weighted MR volumes from 10 subjects. We also implemented equivalent pipelines in Nipype and Bash and measured the execution times of all implementations after deleting (i) all and (ii) half of the intermediate and output files of each pipeline from a first pass. BRICpipe and Nipype pipelines were parallelized across 6 cores of our SMP Linux processing server.
Our results show that BRICpipe is as flexible as Bash scripts and Nipype, and that BRICpipe’s execution times are within -5% and +10% of Nipype’s MultiCore engine.
DISCUSSION
BRICpipe aims to bridge the gap between command line scripts and established pipeline frameworks by making structured pipeline development more intuitive. Unlike LONI and Nipype, BRICpipe can be applied in other fields, as its design is quite generic.
We encourage the use of pipeline frameworks in clinical research, as they are an efficient way to communicate current needs and problems to Neuroinformatics tool developers.
REFERENCES
[1] Gorgolewski K et al. Front. Neuroinform. 5:13. doi:10.3389/fninf.2011.00013
[2] Dinov I et al. PLoS ONE 5(9):e13070. doi:10.1371/journal.pone.0013070
[3] www.gnu.org
[4] www.yworks.com
In clinical research, neuroimaging pipelines are mainly used to combine the strength of single analysis tools and to accelerate the data analysis. Pipeline frameworks, such as Nipype[1] or LONI[2] (Table 1), should be a researcher’s first choice to implement pipelines. However, clinical researchers often prefer Bash[3] scripts due to their simplicity and because pipeline frameworks still require specialist knowledge for setup and use.
To encourage the use of pipeline frameworks in clinical research, we developed our own open source pipeline framework, BRICpipe. Here, we present BRICpipe’s design and some validation results to show how BRICpipe compares to Nipype and Bash scripts.
DESIGN
Figure 1 shows the typical use-case of BRICpipe: the user has to supply a workflow graph consisting of input, output and processing nodes (e.g. Figure 2), a spreadsheet with paths to input data from all subjects, and command line applications for each processing node. The workflow graph is designed in a graph editor (e.g. yEd[4]) and converted to a Makefile with the Workflow2Makefile application. Lastly, the ExecWorkflow application starts the pipeline execution by invoking GNU make[3] as execution engine.
VALIDATION
We validated BRICpipe with a two stage registration pipeline (Figure 2) for clinical T1- and T2*-weighted MR volumes from 10 subjects. We also implemented equivalent pipelines in Nipype and Bash and measured the execution times of all implementations after deleting (i) all and (ii) half of the intermediate and output files of each pipeline from a first pass. BRICpipe and Nipype pipelines were parallelized across 6 cores of our SMP Linux processing server.
Our results show that BRICpipe is as flexible as Bash scripts and Nipype, and that BRICpipe’s execution times are within -5% and +10% of Nipype’s MultiCore engine.
DISCUSSION
BRICpipe aims to bridge the gap between command line scripts and established pipeline frameworks by making structured pipeline development more intuitive. Unlike LONI and Nipype, BRICpipe can be applied in other fields, as its design is quite generic.
We encourage the use of pipeline frameworks in clinical research, as they are an efficient way to communicate current needs and problems to Neuroinformatics tool developers.
REFERENCES
[1] Gorgolewski K et al. Front. Neuroinform. 5:13. doi:10.3389/fninf.2011.00013
[2] Dinov I et al. PLoS ONE 5(9):e13070. doi:10.1371/journal.pone.0013070
[3] www.gnu.org
[4] www.yworks.com
Preferred presentation format:
Poster
Topic:
Clinical neuroscience

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
