Mindboggle 2: Automated human brain MRI feature extraction, identification, shape analysis, and labeling
Filed under:
General neuroinformatics
Arno Klein (Columbia University), Forrest Bao (Texas Tech University), Eliezer Stavsky (Columbia University), Yrjö Häme (Columbia University), Joachim Giard (Universite Catholique de Louvain), Nolan Nichols (Washington University), Satrajit Ghosh (MIT)
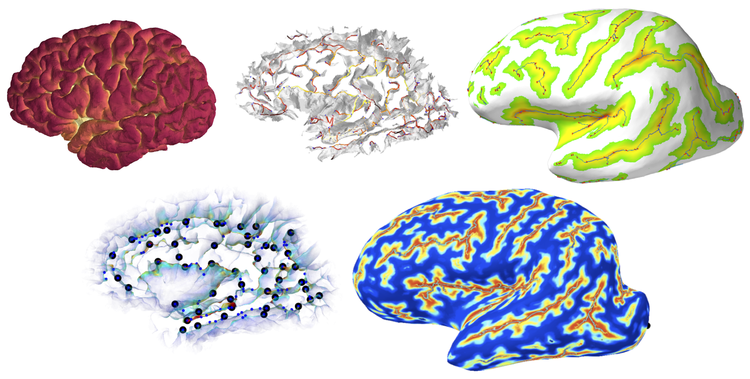
Mindboggle 2 is a new neuroinformatics platform that (1) extracts multiple, nested features from depth and curvature maps of a cortical surface, uses label propagation to (2) segment and identify the features and (3) label the cortical surface in areas between these features, and (4) quantifies the shapes of the identified features and labeled regions. Mindboggle is open source, Python software (http://www.mindboggle.info). We are currently applying Mindboggle in morphometry studies and region-based functional and diffusion MRI analyses.
Features
Mindboggle uses the Nipype pipeline framework to provide a flexible and modular way to include multiple methods for extracting features, including sulcus folds, pits (bottommost points), fundi (curves along the depths of folds), and medial surfaces (“midlines” within folds). For example, one of our pit extraction algorithms assigns a likelihood value to each point based on its depth and local surface curvature, and employs a hidden Markov measure field model to discourage spatially clustered configurations of pit points. We use a similar approach for one of our fundus extraction algorithms, where the probabilistic model is formulated to encourage elongated, connected structures that reach the full length of folds. Our medial surfaces “grow” from our fundi at the depths of a sulcus and are guided upward by vertices belonging to opposite sulcal banks. To identify features, we segment them by distinct pairs of surrounding anatomical labels. We first register multiple, manually labeled brains to a target brain, then propagate labels along the resulting probabilistic label map from consensus labels to the features. For shape analysis, we compute geometric and spectral shape measures for each feature, and sulcus spans using the normals to each medial surface point.
Labels
The above fundi provide a much more consistent means of defining some label boundaries than a human would be capable of. We enforce a closer correspondence between label boundaries and fundi by creating a “fundus friendly” version of the protocol by aggregating regions whose divisions are not defined by fundi, and by post-processing labeled surfaces to conform to the protocol. The label propagation used to identify fundi also automates labeling of the areas between the fundi. The result is therefore a feature-defined, fundus-friendly labeling protocol and an automated means to apply this protocol by moving label boundaries to coincide with fundi.
Features
Mindboggle uses the Nipype pipeline framework to provide a flexible and modular way to include multiple methods for extracting features, including sulcus folds, pits (bottommost points), fundi (curves along the depths of folds), and medial surfaces (“midlines” within folds). For example, one of our pit extraction algorithms assigns a likelihood value to each point based on its depth and local surface curvature, and employs a hidden Markov measure field model to discourage spatially clustered configurations of pit points. We use a similar approach for one of our fundus extraction algorithms, where the probabilistic model is formulated to encourage elongated, connected structures that reach the full length of folds. Our medial surfaces “grow” from our fundi at the depths of a sulcus and are guided upward by vertices belonging to opposite sulcal banks. To identify features, we segment them by distinct pairs of surrounding anatomical labels. We first register multiple, manually labeled brains to a target brain, then propagate labels along the resulting probabilistic label map from consensus labels to the features. For shape analysis, we compute geometric and spectral shape measures for each feature, and sulcus spans using the normals to each medial surface point.
Labels
The above fundi provide a much more consistent means of defining some label boundaries than a human would be capable of. We enforce a closer correspondence between label boundaries and fundi by creating a “fundus friendly” version of the protocol by aggregating regions whose divisions are not defined by fundi, and by post-processing labeled surfaces to conform to the protocol. The label propagation used to identify fundi also automates labeling of the areas between the fundi. The result is therefore a feature-defined, fundus-friendly labeling protocol and an automated means to apply this protocol by moving label boundaries to coincide with fundi.
Preferred presentation format:
Demo
Why demo:
This framework describes many stages of brain image processing, from feature extraction to brain labeling. Rather than submit multiple abstracts, I thought it better to consolidate the descriptions of the multiple stages into one cohesive demonstration that I can tailor to a given group of visitors. And as the data lend themselves to visual representation, this work really requires a visual demonstration to be effective.
Topic:
General neuroinformatics

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
