Neuroimaging in the Browser using the X Toolkit
Filed under:
Neuroimaging
Daniel Haehn (Children's Hospital Boston, Harvard Medical School), Nicolas Rannou (Children's Hospital Boston, Harvard Medical School), Banu Ahtam (Children's Hospital Boston, Harvard Medical School), Ellen Grant (Children's Hospital Boston, Harvard Medical School), Rudolph Pienaar (Children's Hospital Boston, Harvard Medical School)
WebGL is recent technology that exposes a computer’s GPU to a browser, and allows for the native generation of rich three dimensional graphics [1]. Compatible web browsers can offer a graphical experience comparable to more traditional stand-alone programs. As such WebGL offers the potential of bringing the “web” to neuroscience, and has great potential to accelerate and support scientific research.
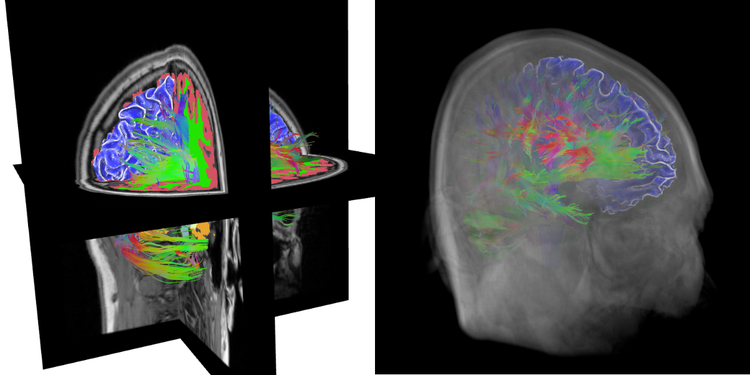
Cognisant of these possibilities, we present ‘The X Toolkit’ (XTK), the first JavaScript-based framework for visualizing and interacting with medical imaging data using WebGL. The toolkit is geared towards powerful scientific visualization and provides a simple API (a 3D DICOM volume can be fully manipulated with three lines of code).
Previous neuroimaging visualization using WebGL has been focused on specific data sets and is not easily generalizable [2],[3]. On the other hand, several frameworks for general WebGL development exist [4], but require a deep understanding of low-level computer graphics programming. Inspired by both approaches, we designed the XTK API to hide low-level elements of WebGL from users and offer native support of established neuroimaging file formats, e.g. supported files can be placed onto a web server and instantly rendered. XTK supports: surface meshes in the Visualization Toolkit, Standard Tessellation, and Freesurfer formats; single-file DICOM volumes and label maps in NRRD and Freesurfer formats; tractography files in the Diffusion Toolkit format; as well as curvature overlays.
XTK is optimized to enable fast data processing and high rendering frame rates. The engine uses caching and optimized loops as well as asynchronous loading. The Google Closure compiler is connected to a CDash infrastructure to monitor the build process and track tests on different browsers. XTK, the build and test system, and all libraries are open sourced and hosted at GitHub [5].
[1] WebGL Specification, Khronos Group 2012, http://www.khronos.org/webgl
[2] Ginsburg D. et al., Realtime Visualization of the Connectome in the Browser using WebGL. 4th INCF Congress of Neuroinformatics 2011. doi: 10.3389/conf.fninf.2011.08.00095
[3] Kelc R., Zygote Body: A New Interactive 3-Dimensional Didactical Tool for Teaching Anatomy. WebmedCentral ANATOMY 2012;3(1):WMC002903
[4] WebGL Frameworks, Khronos Group 2012, http://www.khronos.org/[…]/User_Contributions#Frameworks
[5] XTK, https://github.com/xtk
[6] XTK Visualization, http://demos.goXTK.com/teenager/
Cognisant of these possibilities, we present ‘The X Toolkit’ (XTK), the first JavaScript-based framework for visualizing and interacting with medical imaging data using WebGL. The toolkit is geared towards powerful scientific visualization and provides a simple API (a 3D DICOM volume can be fully manipulated with three lines of code).
Previous neuroimaging visualization using WebGL has been focused on specific data sets and is not easily generalizable [2],[3]. On the other hand, several frameworks for general WebGL development exist [4], but require a deep understanding of low-level computer graphics programming. Inspired by both approaches, we designed the XTK API to hide low-level elements of WebGL from users and offer native support of established neuroimaging file formats, e.g. supported files can be placed onto a web server and instantly rendered. XTK supports: surface meshes in the Visualization Toolkit, Standard Tessellation, and Freesurfer formats; single-file DICOM volumes and label maps in NRRD and Freesurfer formats; tractography files in the Diffusion Toolkit format; as well as curvature overlays.
XTK is optimized to enable fast data processing and high rendering frame rates. The engine uses caching and optimized loops as well as asynchronous loading. The Google Closure compiler is connected to a CDash infrastructure to monitor the build process and track tests on different browsers. XTK, the build and test system, and all libraries are open sourced and hosted at GitHub [5].
[1] WebGL Specification, Khronos Group 2012, http://www.khronos.org/webgl
[2] Ginsburg D. et al., Realtime Visualization of the Connectome in the Browser using WebGL. 4th INCF Congress of Neuroinformatics 2011. doi: 10.3389/conf.fninf.2011.08.00095
[3] Kelc R., Zygote Body: A New Interactive 3-Dimensional Didactical Tool for Teaching Anatomy. WebmedCentral ANATOMY 2012;3(1):WMC002903
[4] WebGL Frameworks, Khronos Group 2012, http://www.khronos.org/[…]/User_Contributions#Frameworks
[5] XTK, https://github.com/xtk
[6] XTK Visualization, http://demos.goXTK.com/teenager/
Preferred presentation format:
Demo
Why demo:
We can present the WebGL developments live in a default browser to show the visualization capabilities and rendering performance.
Topic:
Neuroimaging

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
