Neurons to Algorithms neural circuit model development platform
Filed under:
Large scale modeling
Derek Trumbo (Sandia National Laboratories ), Christina Warrender (Sandia National Laboratories ), James Aimone (Sandia National Laboratories ), Corinne Teeter (Sandia National Laboratories ), Fred Rothganger (Sandia National Laboratories )
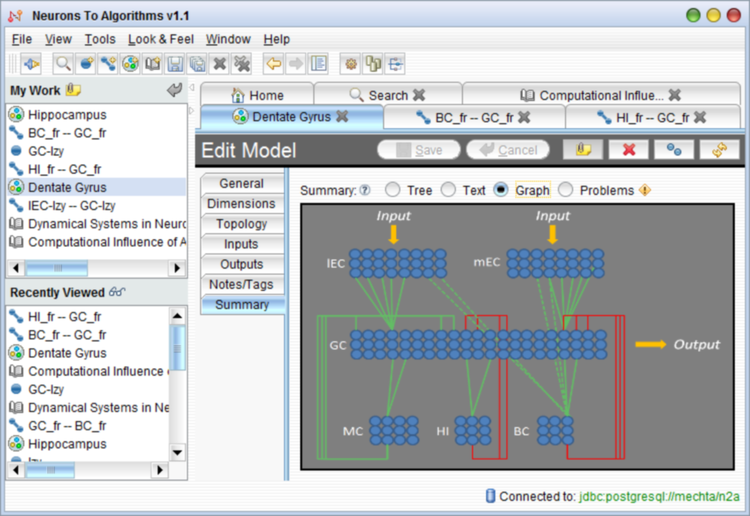
We are developing a software package “Neurons to Algorithms”, or “N2A”. The scientific purpose of N2A is to facilitate biologically realistic neural network model development by 1) compiling (and referencing) neural data from many users and sources in a computable format, 2) automating the model development process by constructing models specific to an individual user’s research goals, and 3) integrating the process by which a model is simulated and analyzed.
A priority in the system’s design is maximizing both the utility and usability for neuroscientists. In contrast to many scientific tools with open source intentions, we have focused considerable effort towards user interface. Users are able to generate models at multiple levels of resolution (spiking or firing rate; point neuron or morphological; networks or individual neurons). The tool has been designed to operate in either a local mode, where the user creates their own database of biological information and details necessary for their modeling, or a distributed mode where users can create models from a community database. Our vision is that this community database will enable the generation and sharing of high fidelity model components across laboratories.
In order to take full advantage of the rapidly expanding neural data sets available, N2A will directly interface the modeling database (in which neuroscience data is represented in a computable format) with existing neuroinformatics databases on anatomy, gene expression, whose data is typically represented in a more conventional format. Furthermore, we plan to integrate N2A with existing simulation platforms that are widely used in the community for specific research questions (e.g., Neuron).
We will demonstrate the N2A platform using a model of the hippocampus (Aimone et al, 2009).
A priority in the system’s design is maximizing both the utility and usability for neuroscientists. In contrast to many scientific tools with open source intentions, we have focused considerable effort towards user interface. Users are able to generate models at multiple levels of resolution (spiking or firing rate; point neuron or morphological; networks or individual neurons). The tool has been designed to operate in either a local mode, where the user creates their own database of biological information and details necessary for their modeling, or a distributed mode where users can create models from a community database. Our vision is that this community database will enable the generation and sharing of high fidelity model components across laboratories.
In order to take full advantage of the rapidly expanding neural data sets available, N2A will directly interface the modeling database (in which neuroscience data is represented in a computable format) with existing neuroinformatics databases on anatomy, gene expression, whose data is typically represented in a more conventional format. Furthermore, we plan to integrate N2A with existing simulation platforms that are widely used in the community for specific research questions (e.g., Neuron).
We will demonstrate the N2A platform using a model of the hippocampus (Aimone et al, 2009).
Preferred presentation format:
Demo
Why demo:
We have developed very user friendly neural network software platform. It would be very useful to be able to demonstrate it's functionality. Also we plan to allow people to create their own models on our computer at the conference.
Topic:
Large scale modeling

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
