Simulator for Realistic High-Density Microelectrode Array Signals
Filed under:
General neuroinformatics
David Jäckel (ETH Zurich, Department of Biosystems Science and Engineering), Urs Frey (RIKEN, Quantitative Biology Center), Felix Franke (ETH Zurich, Department of Biosystems Science and Engineering), Andreas Hierlemann (ETH Zurich, Department of Biosystems Science and Engineering)
High-density microelectrode arrays (HD-MEAs), which allow for performing extracellular recordings at subcellular resolution, have become an important means to study the information processing of neuronal networks. The large number of recording electrodes and the highly redundant nature of HD-MEA data, however, pose challenges to the analysis so that appropriate strategies and novel spike-sorting techniques have to be developed. In order to evaluate the performance of spike sorting techniques for HD-MEA recordings, simulated data can be used, for which the spike trains of the neurons are known.
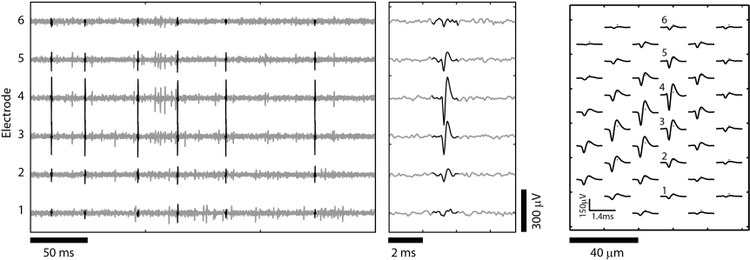
Here we present a toolbox for simulating realistic neuronal HD-MEA signals, implemented in Matlab. The simulator uses a library of well-isolated templates (model templates), which were extracted from HD-MEA recordings using manually supervised spike sorting (see Figure 1). The spatial resolution of the model templates is increased by interpolating on a grid (typically 5 µm pitch). By modifying orientation, amplitude and spatial extension of the model templates, individual neuronal templates are simulated. The modified templates can be positioned over the array, allowing for simulating a wide range of neuronal arrangements. The positioning of the electrodes is arbitrarily set by the user, which allows evaluating the spike sorting performance for different electrode configurations.
Individual spike trains are assigned to the simulated neurons. A noise signal, recorded under experimental conditions with a preparation on the array that had no visible spiking activity, is then added to the simulated spike data. Additionally, neuronal background noise can be simulated adding numerous neurons spiking with low amplitudes. The resulting signal is quantified to a least significant bit as the one used in the measurements.
The presented simulator for neuronal high-density MEA recordings has been used in [1] to evaluate the performance of independent-component-analysis-based spike sorting for HD-MEA retinal recordings. It was also used to simulate activity of Purkinje cells in cerebellar brain slices, and can be used for other biological preparations.
[1] D. Jackel, U. Frey, M. Fiscella, F. Franke, and A. Hierlemann, “Applicability of Independent Component Analysis on High-Density Microelectrode Array Recordings,” J Neurophysiol, p. jn.01106.2011-, Apr. 2012.
Here we present a toolbox for simulating realistic neuronal HD-MEA signals, implemented in Matlab. The simulator uses a library of well-isolated templates (model templates), which were extracted from HD-MEA recordings using manually supervised spike sorting (see Figure 1). The spatial resolution of the model templates is increased by interpolating on a grid (typically 5 µm pitch). By modifying orientation, amplitude and spatial extension of the model templates, individual neuronal templates are simulated. The modified templates can be positioned over the array, allowing for simulating a wide range of neuronal arrangements. The positioning of the electrodes is arbitrarily set by the user, which allows evaluating the spike sorting performance for different electrode configurations.
Individual spike trains are assigned to the simulated neurons. A noise signal, recorded under experimental conditions with a preparation on the array that had no visible spiking activity, is then added to the simulated spike data. Additionally, neuronal background noise can be simulated adding numerous neurons spiking with low amplitudes. The resulting signal is quantified to a least significant bit as the one used in the measurements.
The presented simulator for neuronal high-density MEA recordings has been used in [1] to evaluate the performance of independent-component-analysis-based spike sorting for HD-MEA retinal recordings. It was also used to simulate activity of Purkinje cells in cerebellar brain slices, and can be used for other biological preparations.
[1] D. Jackel, U. Frey, M. Fiscella, F. Franke, and A. Hierlemann, “Applicability of Independent Component Analysis on High-Density Microelectrode Array Recordings,” J Neurophysiol, p. jn.01106.2011-, Apr. 2012.
Preferred presentation format:
Poster
Topic:
General neuroinformatics

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
