The NeuroML C. elegans Connectome
Filed under:
Computational neuroscience
Tim Busbice (Interintelligence Research), Padraig Gleeson (University College London), Sergey Khayrulin (A.P. Ershov Institute of Informatics Systems), Matteo Cantarelli (OpenWorm.org), Alexander Dibert (A.P. Ershov Institute of Informatics Systems), Giovanni Idili (OpenWorm.org), Andrey Palyanov (A.P. Ershov Institute of Informatics Systems), Stephen Larson (OpenWorm.org)
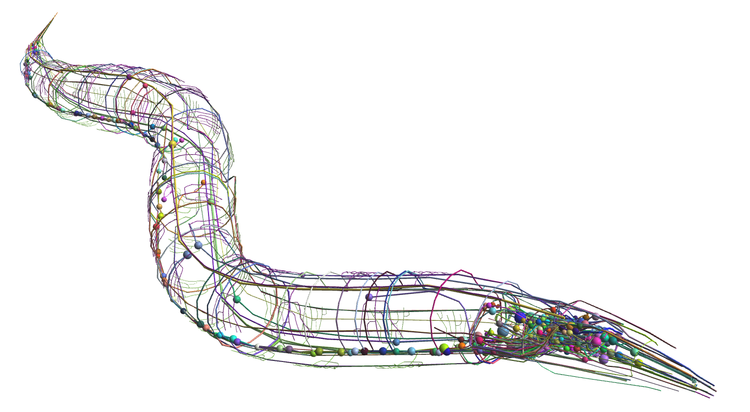
We have merged and extended the C. elegans connectome (Varshney et al., 2006) and a three-dimensional cellular anatomy model (Grove & Sternberg, 2011) in the context of the OpenWorm project, an open source project to build a data integration and simulation framework for the C. elegans. To do so, we have leveraged the NeuroML standard (Gleeson et al., 2010), a language for describing neuronal morphologies, ion channels, synapse models and position and network structure in a simulator independent format. It facilitates the exchange of these key model components between computational neuroscience applications. We have converted the neurons described as 3D objects into NeuroML multi-compartmental neuron models and populated the connection statements between these neurons with the Varshney et al. (2006) connection graph. We have used NeuroConstruct (Gleeson et al., 2007) as the rallying point for these data integration efforts and we have demonstrated a successful export from NeuroConstruct into a simulation engine. We have also made available a WebGL based browser that enables the neurons to be seen in the 3D context of the rest of the C. elegans anatomy (http://browser.openworm.org). While not yet sufficient to explain the activity of its neurons, we believe rthis is a necessary prerequisite for deep investigations into the non-linear dynamics and neuronal computation of the C. elegans neuronal network.
Supported by the Neurolinx Research Institute (http://neurolinx.org)
References
Gleeson, P., Steuber, V., & Silver, R. A. (2007). neuroConstruct: a tool for modeling networks of neurons in 3D space. Neuron, 54(2), 219-35. http://dx.doi.org/10.1016/j.neuron.2007.03.025
Gleeson, P., Crook, S., Cannon, R. C., Hines, M. L., Billings, G. O., Farinella, M., Morse, T. M., et al. (2010). NeuroML: A Language for Describing Data Driven Models of Neurons and Networks with a High Degree of Biological Detail. (K. J. Friston, Ed.)PLoS Computational Biology, 6(6), e1000815. http://dx.doi.org/10.1371/journal.pcbi.1000815
Grove, C.A., Sternberg, P.W.. (2011) The Virtual Worm: A Three-Dimensional Model of the Anatomy of Caenorhabditis elegans at Cellular Resolution. 18th International C. elegans meeting. Retrieved from http://www.celegans.org/2011/pdf/abstracts_web.pdf
Varshney, L. R., Chen, B. L., Paniagua, E., Hall, D. H., & Chklovskii, D. B. (2009). Structural properties of the caenorhabditis elegans neuronal network. Arxiv preprint arXiv:0907.2373, 12596, 1-41.
Supported by the Neurolinx Research Institute (http://neurolinx.org)
References
Gleeson, P., Steuber, V., & Silver, R. A. (2007). neuroConstruct: a tool for modeling networks of neurons in 3D space. Neuron, 54(2), 219-35. http://dx.doi.org/10.1016/j.neuron.2007.03.025
Gleeson, P., Crook, S., Cannon, R. C., Hines, M. L., Billings, G. O., Farinella, M., Morse, T. M., et al. (2010). NeuroML: A Language for Describing Data Driven Models of Neurons and Networks with a High Degree of Biological Detail. (K. J. Friston, Ed.)PLoS Computational Biology, 6(6), e1000815. http://dx.doi.org/10.1371/journal.pcbi.1000815
Grove, C.A., Sternberg, P.W.. (2011) The Virtual Worm: A Three-Dimensional Model of the Anatomy of Caenorhabditis elegans at Cellular Resolution. 18th International C. elegans meeting. Retrieved from http://www.celegans.org/2011/pdf/abstracts_web.pdf
Varshney, L. R., Chen, B. L., Paniagua, E., Hall, D. H., & Chklovskii, D. B. (2009). Structural properties of the caenorhabditis elegans neuronal network. Arxiv preprint arXiv:0907.2373, 12596, 1-41.
Preferred presentation format:
Poster
Topic:
Computational neuroscience

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
