TissueStack: a new way to view your imaging data
Filed under:
General neuroinformatics
Andrew Janke (Center for Advanced Imaging), Harald Waxenegger (Center for Advanced Imaging), Jeremy Ullmann (Center for Advanced Imaging), Graham Galloway (Center for Advanced Imaging)
It's no secret that the increases in resolution possible with modern imaging equipment has led to an explosion in data. This increase is not always something that is easy to deal with, especially in studies that involve multiple modalities and sites. University and research networks while continually improving still do not allow those in the neuroinformatics field to seamlessly share multi-terrabye images.
This leads to the problem in multi-site studies and often in smaller projects in which no one is sure where or how to access the current version of all the data without having to download TB's of data, make a small change and then upload again. This problem is not unique to the neuroimaging field and as such we have endeavoured to make use of techniques from the very closely related field of GIS (Graphical Information Systems) in this project.
There has been some work done in this area, most notably with the web interfaces of the Allen Brain atlas and CATMAID. Both of these however are primarilly written for the viewing of multiple 2D images, typically of histology.
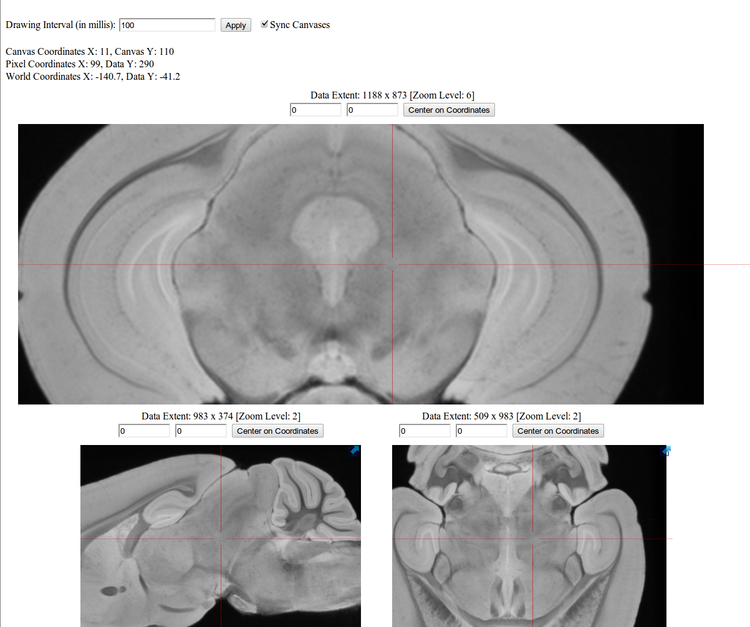
TissueStack is an Open Source project that is currently available on GitHub (http://github.com/NIF-au/TissueStack) and at the time of this abstract is in alpha release phase for comment. Its purpose is to allow researchers to view multi-TB imaging datasets online vai an interface style that most people are familiar with (online mapping) at reasonable speed. The application uses HTML5 Canvas elements and thus will work equally well on mobile devices (tablets, phones, etc). The current proof of concept can be viewed at http://www.imaging.org.au/tissuestack and is displaying a 850MB 30um c57/bl mouse model.
The application generates multi-scale tiled images from an input dataset in order to optimise viewing speed for a given network connection. Data input formats currently include MINC, NiFTI and OpenSlide formats. Future versions of the application will allow the federation of data from multiple sites and the overlay of pre-aligned muti-modality data such Histology and MRI.
Acknowlegements: This project is supported by the Australian National Data Service (ANDS). ANDS is supported by the Australian Government through the National Collaborative Research Infrastructure Strategy Program and the Education Investment Fund (EIF) Super Science Initiative.
This leads to the problem in multi-site studies and often in smaller projects in which no one is sure where or how to access the current version of all the data without having to download TB's of data, make a small change and then upload again. This problem is not unique to the neuroimaging field and as such we have endeavoured to make use of techniques from the very closely related field of GIS (Graphical Information Systems) in this project.
There has been some work done in this area, most notably with the web interfaces of the Allen Brain atlas and CATMAID. Both of these however are primarilly written for the viewing of multiple 2D images, typically of histology.
TissueStack is an Open Source project that is currently available on GitHub (http://github.com/NIF-au/TissueStack) and at the time of this abstract is in alpha release phase for comment. Its purpose is to allow researchers to view multi-TB imaging datasets online vai an interface style that most people are familiar with (online mapping) at reasonable speed. The application uses HTML5 Canvas elements and thus will work equally well on mobile devices (tablets, phones, etc). The current proof of concept can be viewed at http://www.imaging.org.au/tissuestack and is displaying a 850MB 30um c57/bl mouse model.
The application generates multi-scale tiled images from an input dataset in order to optimise viewing speed for a given network connection. Data input formats currently include MINC, NiFTI and OpenSlide formats. Future versions of the application will allow the federation of data from multiple sites and the overlay of pre-aligned muti-modality data such Histology and MRI.
Acknowlegements: This project is supported by the Australian National Data Service (ANDS). ANDS is supported by the Australian Government through the National Collaborative Research Infrastructure Strategy Program and the Education Investment Fund (EIF) Super Science Initiative.
Preferred presentation format:
Poster
Topic:
General neuroinformatics

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
