Virtual Fly Brain - a data hub for Drosophila neurobiology
Filed under:
Digital atlasing
David Osumi-Sutherland (University of Cambridge), Nestor Milyaev (Edinburgh University), Marta Costa (University of Cambridge), Gregory S.X.E. Jefferis (MRC Laboratory of Molecular Biology), Cahir J. O'Kane (University of Cambridge), J. Douglas Armstrong (Edinburgh University)
Drosophila neurobiology is on the cusp of a data explosion that could facilitate significant advances in our understanding of basic neurobiology. Realizing this promise will require intuitive tools that allow queries across diverse data types from multiple sources, including the literature. Virtual Fly Brain (VFB) [1] already partially fulfills this role, integrating neuroanatomical data from the literature with genomic and genetic data in the FlyBase genetic database. We are beginning to integrate bulk data, including large sets of annotated 3D images.
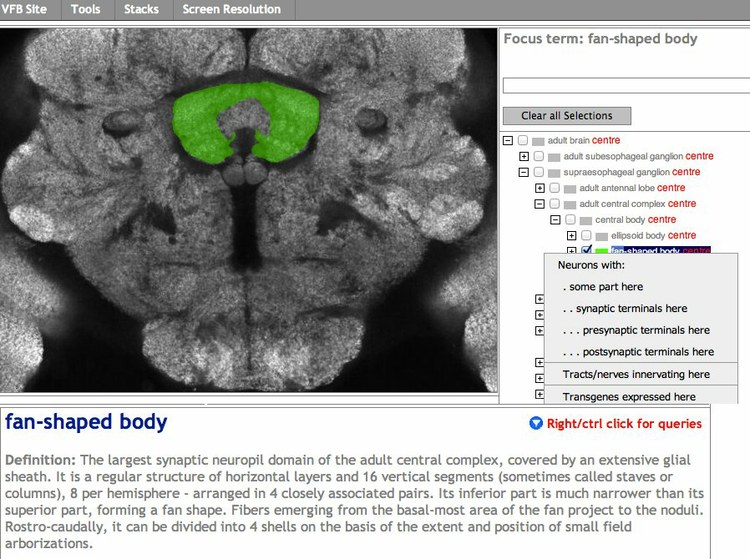
An extensive ontology of Drosophila neuroanatomy provides the glue for data integration and the substrate for queries on VFB. This ontology is used by FlyBase to annotate expression and phenotype via a simple tagging model, but it also allows more sophisticated forms of data integration. The ontology uses the W3C standard ontology language OWL2 and a set of general relations for representing neuroanatomy [2], developed in coordination with the INCF funded Project for Ontologies in Neuroscience. The basic spatial reasoning that these relations allow is vital to VFB. It drives our queries of brain regions for innervating neurons, expression patterns and phenotypes and will soon drive our queries of annotated neuron images. Where individual annotated neurons can be mapped to known neuron classes, we can use information about the mapped class to enrich image queries. Conversely, we can use information extracted from neuron images about the location of neuron parts to enrich queryable information about mapped neuron classes. Our ontology also includes extensive use of relations for recording lineage, neurotransmitter and function and synaptic connections. The VFB query system will soon be extended to encompass these.
We work with data providers to annotate images in a form that we can easily integrate. Where this is not possible, we analyse bulk image data using a pipeline that registers images to a standard, extracts spatial information and clusters neurons by shape. Where clustering predicts new isomorphic neuron classes, we incorporate these into our ontology.
As well as providing a data integration hub for Drosophila neurobiology, our system has great potential for generalisation to other systems in neurobiology.
[1] http://www.virtualflybrain.org; Milyaev et al., 2012 http://dx.doi.org/10.1093/bioinformatics/btr677
[2] Osumi-Sutherland et al., 2012 http://dx.doi.org/10.1093/bioinformatics/bts113
An extensive ontology of Drosophila neuroanatomy provides the glue for data integration and the substrate for queries on VFB. This ontology is used by FlyBase to annotate expression and phenotype via a simple tagging model, but it also allows more sophisticated forms of data integration. The ontology uses the W3C standard ontology language OWL2 and a set of general relations for representing neuroanatomy [2], developed in coordination with the INCF funded Project for Ontologies in Neuroscience. The basic spatial reasoning that these relations allow is vital to VFB. It drives our queries of brain regions for innervating neurons, expression patterns and phenotypes and will soon drive our queries of annotated neuron images. Where individual annotated neurons can be mapped to known neuron classes, we can use information about the mapped class to enrich image queries. Conversely, we can use information extracted from neuron images about the location of neuron parts to enrich queryable information about mapped neuron classes. Our ontology also includes extensive use of relations for recording lineage, neurotransmitter and function and synaptic connections. The VFB query system will soon be extended to encompass these.
We work with data providers to annotate images in a form that we can easily integrate. Where this is not possible, we analyse bulk image data using a pipeline that registers images to a standard, extracts spatial information and clusters neurons by shape. Where clustering predicts new isomorphic neuron classes, we incorporate these into our ontology.
As well as providing a data integration hub for Drosophila neurobiology, our system has great potential for generalisation to other systems in neurobiology.
[1] http://www.virtualflybrain.org; Milyaev et al., 2012 http://dx.doi.org/10.1093/bioinformatics/btr677
[2] Osumi-Sutherland et al., 2012 http://dx.doi.org/10.1093/bioinformatics/bts113
Preferred presentation format:
Demo
Why demo:
A demo is required to get the full possibilities for data integration using our system across.
Topic:
Digital atlasing

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
